Adenylyl Cyclase
This web page was produced as an assignment for an undergraduate course at Davidson College.
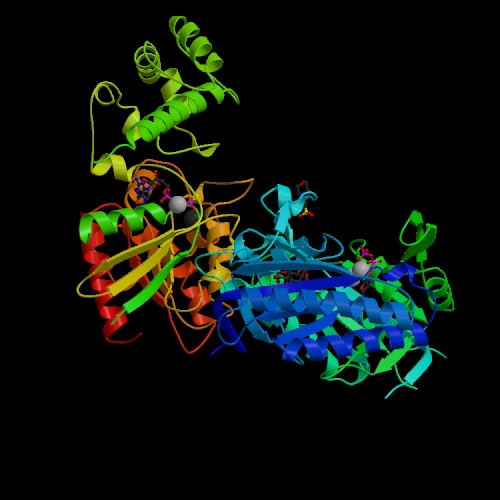
Figure 1. Image of adenylyl cyclase. This image, adenylyl cyclase bound to a signaling protein, reveals that the adenylyl cyclase protein is made up of both α-helices and β-sheets. Image obtained from PDB (PDB, 2005).
Introduction
Adenylyl cyclase (AC) is an enzyme, with nine cloned mammalian isoforms, located in the plasma membrane of a cell that is important in the regulation of many cellular signals and pathways. AC catalyzes the transformation of ATP to cyclic AMP (cAMP), a second messenger. cAMP is released into the cytoplasm after a first messenger G protein binds to its receptor on the catalytic domains of adenylyl cyclase. Once adenylyl cyclase forms cAMP and releases it into the cytoplasm, the cAMP binds to ion channels or to an enzyme, such as protein kinase, to expose this enzyme's active site, which continues the communication pathway (Purves, 2001 and Bowen, 2000).
Structure
The basic structures of the mammalian adenylyl cyclases have twelve hydrophobic, transmembrane sections, loops that link these sections, and two catalytic domains, the ‘palm’ domain and the adenylyl cyclase catalytic core (ACYc) domain (Zhang, 1997 and Artymiuk, 1997). The 'palm' domain, which is very similar to the structure of DNA polymerase, is made up of four β-strands and three α-helices. The ACYc domain does not have significant differences from the palm domain, but it does lack the fingers regions that the first stand has and form a β-sheet on the lower part of the amino terminus which extends over the analogous thumb region located in the palm domain (see Figure 2) (Artymiuk, 1997).
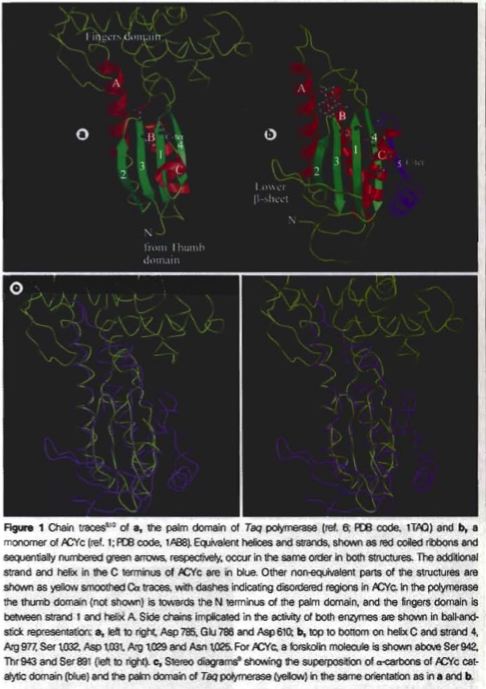
Figure 2. Images of the two catalytic domains of adenylyl cyclase. These images demonstrate the similarities and differences between the two catalytic domains of adenylyl cyclase. Image obtained from Artymiuk, 1997 (permission pending).
All AC enzymes have many stimulatory and inhibitory receptors that bind with the G proteins and moderate the catalytic subunit activity of AC. All AC proteins are able to be activated by the binding of a G protein to its receptor (Hsueh, 2001).

Figure 3. Image of adenylyl cyclase. This image reveals the binding receptor in adenylyl cyclase, indicated by the red region, for the G proteins. Image obtained from http://www.cs.stedwards.edu/chem/Chemistry/CHEM43/CHEM43/GProtein/function.html (permission pending).
The two previously mentioned catalytic domains face each other and form a wreath-like cavity, which has been found to be important in regulator binding for some isoforms of adenylyl cyclase (Artymiuk, 1997). At the end of this cavity, two forskolin molecules bind to form hydrophobic pockets. The center of this cavity is lined with highly conserved, polar and charged residues which infers that there is ATP binding at this site (Zhang, 1997).
Function
The receptor sites on adenylyl cyclase for the G proteins are incredibly important parts of the protein's structure that allow it to function. When the G proteins bind to the receptors, the binding sites for ATP are exposed and the enzyme is then able to catalyze the transformation of ATP to cAMP (Hsueh, 2001). Since there are both inhibitory and stimulatory receptor sites for the G proteins, the binding of these proteins to AC can increase the production of cAMP, but can decrease the production of cAMP as well.
Since the adenylyl cyclase is a transmembrane protein, it can react to events on one side of the plasma membrane and stimulate many other events within the cell. This enzyme, once it forms cAMP, begins signaling cascades within the cell that can activate other kinases that are able to open ion channels or expose the active sites of other proteins (Purves, 2001).
The two catalytic domains of the adenylyl cyclase enzyme and their interactions provide an binding site for ATP that activates the adenylyl cyclase to transform ATP to cAMP. Without the similarity of the structures of these two domains, the hydrophobic regions would not come together to form the pocket for the ATP to bind, producing cAMP to stimulate the communication pathways within the cell (Zhang, 1997).
Even more about adenylyl cyclase...
Possible link between adenylyl cyclase and Alzheimer's disease? Click here for the astract.
Bacterial adenylyl cyclase is a component of a biothreat agent. Which one?
Davidson College Biology Homepage
Davidson College Molecular Biology Homepage
Email with any questions or comments
References:
Artymiuk, P.J., Poirrette, A. R., Rice, D. W., & Willett, P. "A polymerase I palm in adenylyl cyclase?" Nature 388, 33-34 (1997).
Bowen, R. "Adenylyl Cyclase." Adenylyl Cyclase. 3 Aug. 2000. Accessed 14 Feb. 2005 <http://arbl.cvmbs.colostate.edu/hbooks/molecules/cyclase.html>.
"Functions of G-Proteins and Adenylyl Cyclases." Functions of G-Proteins, Adenylyl Cyclases, and Their Interactions. 15 Feb. 2005 <http://www.cs.stedwards.edu/chem/Chemistry/CHEM43/CHEM43/GProtein/function.html>.
Hsueh, Aaron J. "Adenylate Cyclase 5." Gene Information. 29 Aug. 2001. Stanford University. Accessed 14 Feb. 2005 <http://ovary.stanford.edu/4_display.html?rec=599>.
[PDB] Protein Data Bank. 2004. <http://www.rcsb.org/pdb/index.html>. Accessed 14 Feb 2005. PDB ID: 1CJT.
Purves, William K., David Sadava, Gordon H. Orians, and H. Craig Heller. Life: The Science of Biology. 6th ed. Sunderland: Sinauer Associates, Inc., 2001.
Zhang, G., Liu, Y., Ruoho, A. E. & Hurley, J.H. "Structure of the adenylyl cyclase catalytic core." Nature 386, 247-253 (1997).
© Copyright 2005 Department of Biology, Davidson College, Davidson, NC 28035